Routine metabolomic profiling combining primary metabolism (GC-TOF MS), biogenic amines (HILIC-QTOF MS) and lipidomics (CSH-QTOF MS) yields relative quantitative data for up to 800 identified compounds in typical samples. With these lists of compounds, a range of biological questions can be answered. However, in each project we routinely find many metabolic peaks with unidentified chemical structure that are abundant and statistically significant in case/control studies.
We offer collaborative projects with the Fiehn research laboratory to engage in identification of such unknown peaks, especially for collaborative grant submissions. One aspect of our research is to increase metabolite mass spectral libraries, including acquiring mass spectra for novel metabolite standards with accurate mass MS/MS analysis, accurate mass GC-QTOF MS and accurate mass ion tree fragmentations (MS^n). Another aspect of such research is to improve algorithms for modeling mass spectra from virtual compound libraries, to implement and develop methods for retention time prediction, and to utilize workflows that integrate multiple steps for compound identification.
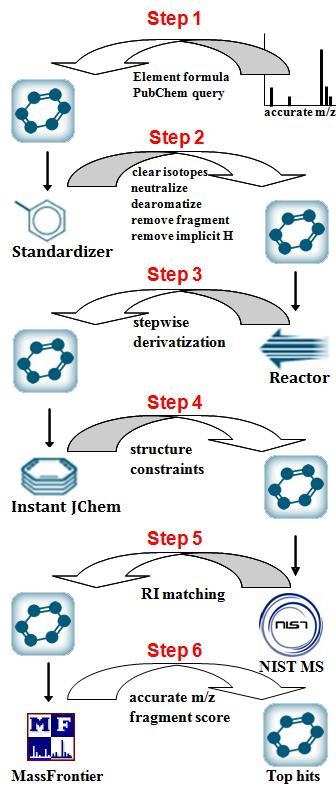
The Experimental Core, led by Prof. Oliver Fiehn generates mass spectral libraries and uploads these to the MassBank of North America. The Core also generates standardized, retention-time locked chromatography methods. The Core uses derivatizations to constrain isomer structures in the data identification workflow (see figure left). Ultimately, these standardized methods are used to develop an automatic LC-MS metabolome annotation and query database similar to the existing BinBase system for GC-MS data.
If interested, please contact Professor Oliver Fiehn (email) for more information.
The administrative core organizes the Center's outreach courses, workshops and seminars.
Please, use our public software, databases and informatics tools!